How to assess cell biology in the context of a tissue
Leveraging spatial multi-omics with Hyperion systems: High-plex imaging of protein and RNA on the same slide
Spatial transcriptomics is a method enabling high-dimensional investigation of gene transcription in a spatial context. By characterizing expression profiles, spatial transcriptomics is important for analyzing transcriptional patterns and regulation as well as identifying cellular neighborhoods and characteristics contributing to disease. Spatial proteomics, on the other hand, is the method of acquiring high-dimensional images of protein expression in a spatial context.
While single-cell transcriptomics (or scRNA-seq) has become a standard in clinical and translational research, the need to preserve intact and viable cells excludes many cell types and destroys any organizational learnings. Spatial transcriptomics addresses these limitations, providing a comprehensive understanding of cell identity and function relative to neighboring cells. Similarly, while regular pathology proteomics (for example, regular DAB IHC) is used to investigate protein expression in tissues, spatial proteomics enables the analysis of 40 or more markers in a single tissue section.
How to assess cell biology in the context of a tissue
Leveraging spatial multi-omics with Hyperion systems: High-plex imaging of protein and RNA on the same slide
Spatial transcriptomics is a method enabling high-dimensional investigation of gene transcription in a spatial context. By characterizing expression profiles, spatial transcriptomics is important for analyzing transcriptional patterns and regulation as well as identifying cellular neighborhoods and characteristics contributing to disease. Spatial proteomics, on the other hand, is the method of acquiring high-dimensional images of protein expression in a spatial context.
While single-cell transcriptomics (or scRNA-seq) has become a standard in clinical and translational research, the need to preserve intact and viable cells excludes many cell types and destroys any organizational learnings. Spatial transcriptomics addresses these limitations, providing a comprehensive understanding of cell identity and function relative to neighboring cells. Similarly, while regular pathology proteomics (for example, regular DAB IHC) is used to investigate protein expression in tissues, spatial proteomics enables the analysis of 40 or more markers in a single tissue section.
Is it possible to look at RNA and proteins in a spatial context at the same time?
The transcriptome and proteome each provide distinct layers of information that support the understanding of biological processes. While mRNA levels show the potential for protein production, protein levels indicate which proteins have been produced and are actionable targets.
A novel workflow combines spatial transcriptomic platforms and the Hyperion™ XTi Imaging System to:
Localize both transcripts and proteins in the same cells
Provide valuable insights into RNA and protein regulation and function in tissue microenvironments
Why integrate spatial transcriptomics and proteomics?
Combining spatial transcriptomics with high-plex proteomic imaging provides a more complete view of tissue biology than either modality alone. Transcriptomic imaging reveals gene expression activity, while proteomics captures functional state – what the cell is actually doing. Because immune cell phenotypes are typically defined by protein expression, not RNA, cell identity can only be inferred from transcripts but is confirmed through protein detection.
Though mRNA and protein levels are related, they often diverge due to regulatory controls, timing delays and biological noise. A fixed tissue section offers only a snapshot in time, and RNA and protein abundances may not align at that moment. By imaging both, researchers capture complementary layers of cell activity.
This dual approach, enabled by sequential use of spatial transcriptomics and Imaging Mass Cytometry™ technology with the Hyperion XTi Imaging System on the same slide, unlocks multidimensional insights into cell states, tissue organization and disease mechanisms. Standard BioTools™ Services Labs support this integrated workflow to help maximize the value of every sample.
How it works: IMC offers flexibility to complement other spatial modalities using the same sample
IMC™ workflows can easily be added after transcriptomic imaging and/or an H&E workflow. The Hyperion XTi workflow directly follows transcriptomic acquisition on the same slide for a critical additional layer of spatial proteomic data. H&E staining can be incorporated between transcriptomic and Hyperion workflows, if desired.
IMC technology stains and then acquires all markers simultaneously. This helps minimize tissue degradation prior to acquisition, greatly reduces assay development requirements and helps make this type of multi-omic workflow not only possible, but routine.
Integration of Hyperion XTi protein imaging on the same slide following RNA imaging
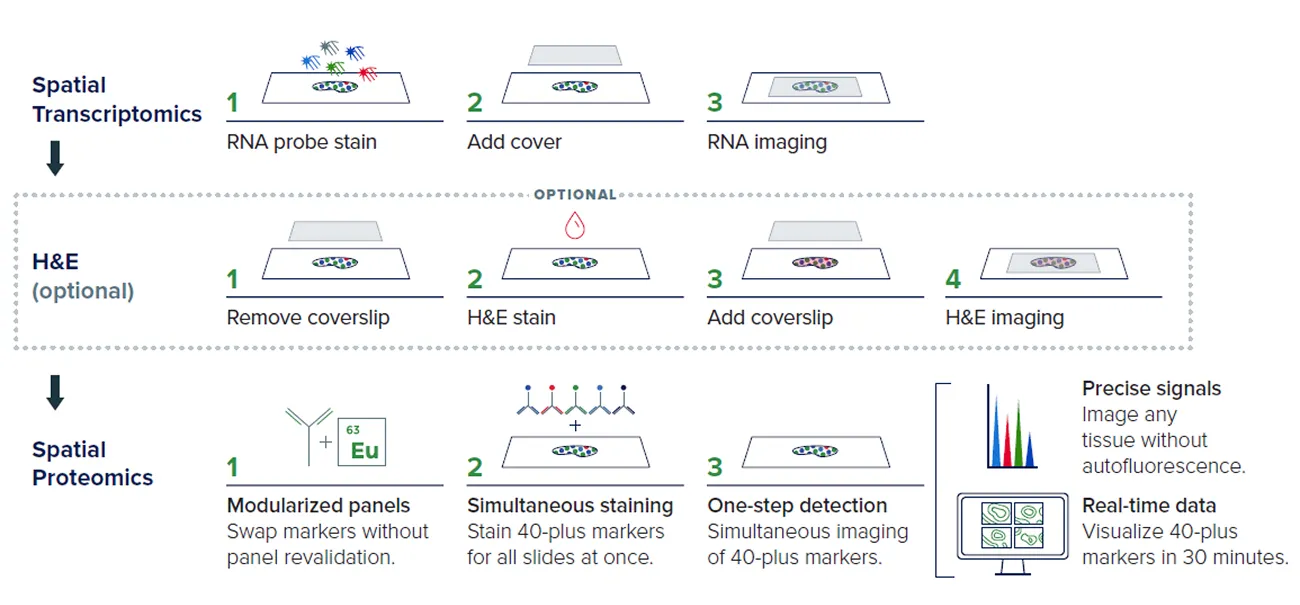
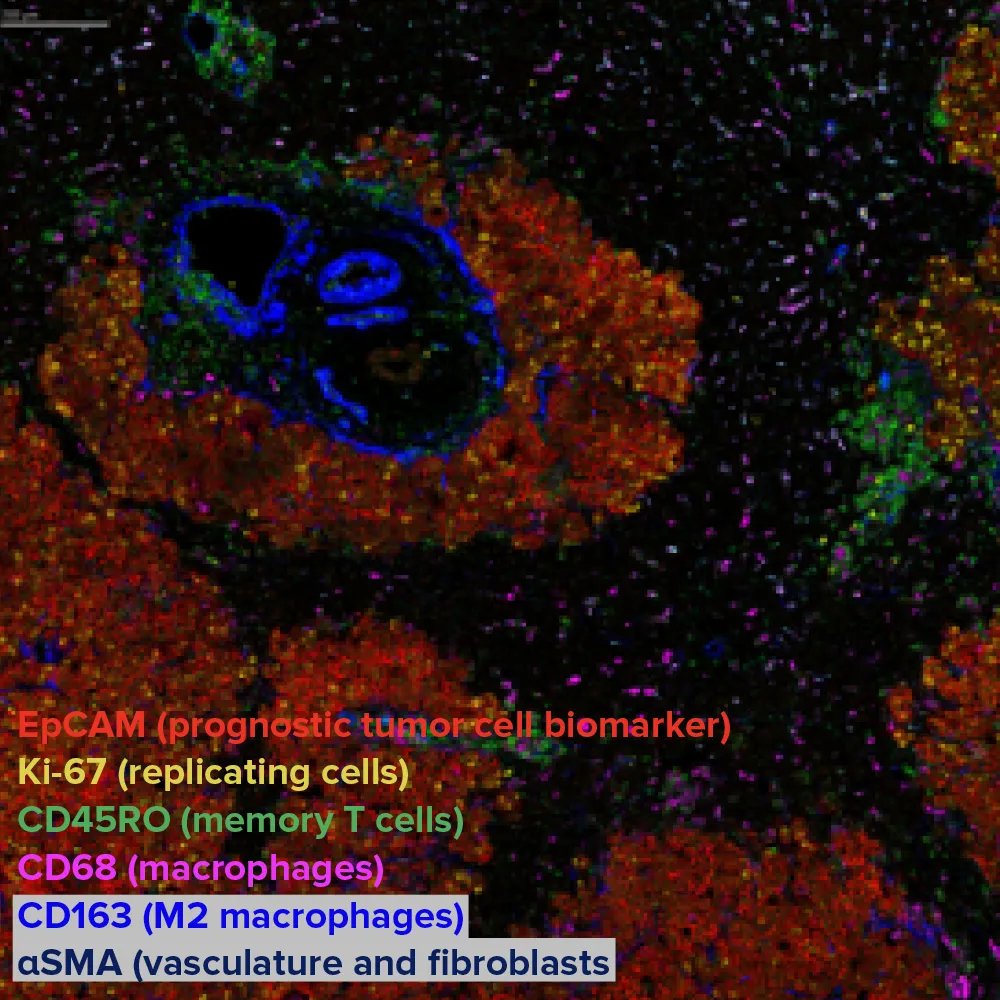
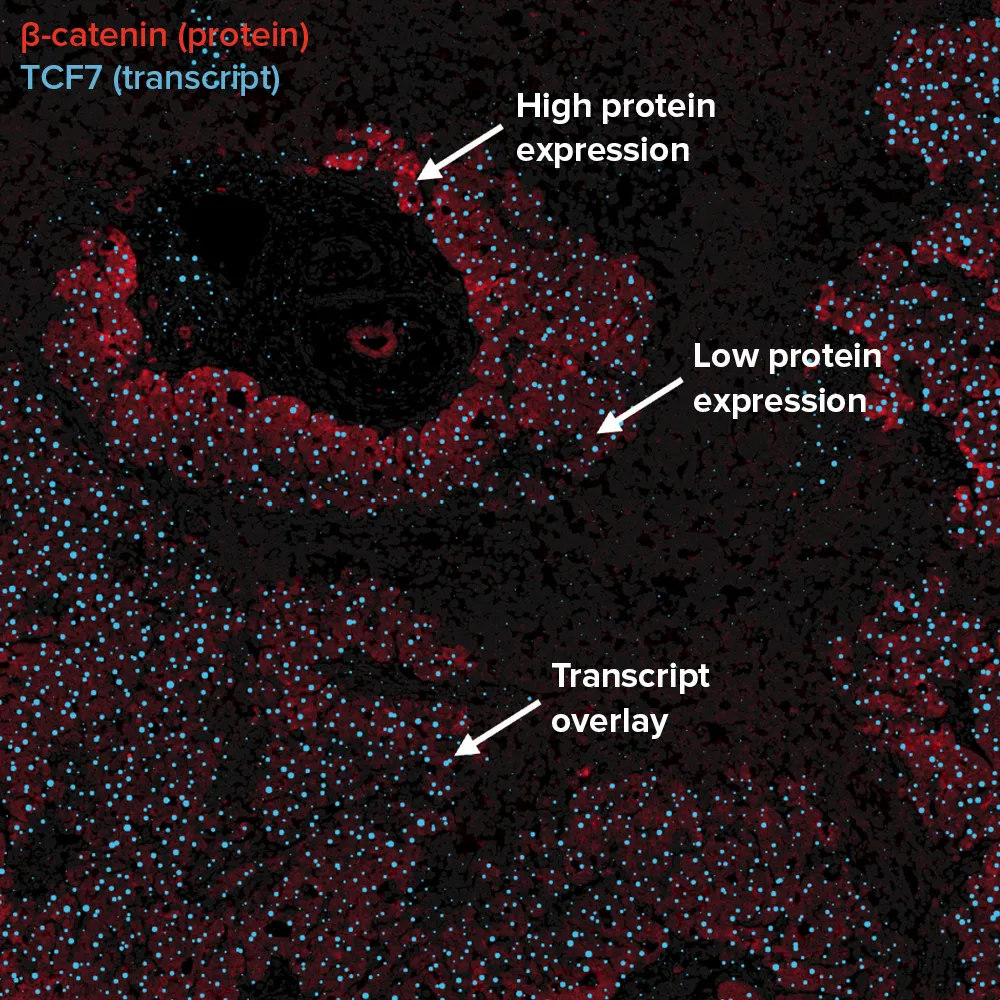
Image of human hepatocellular carcinoma tissue
The same slide can be re-imaged post-transcriptomics using IMC technology, capturing over 40 protein markers in a single multiplexed acquisition.
IMC platforms enable the comparison of spatial transcriptomic data with a choice of up to 45 spatial proteomic markers in the same section. This image shows six of 43 markers detected in the same scan.
- The IMC workflow can follow a spatial transcriptomic workflow on the same slide to deliver high-quality complementary data
- Seamless integration of the two approaches is enabled by simplicity of the IMC workflow and existence of available data analysis solutions
- H&E staining can be incorporated between spatial transcriptomics and IMC application for generating data for pathological review of tissue
- Cell phenotyping capabilities for tumor and immune components of the tissues remain intact in post-transcript detection slides imaged with IMC platforms
Add proteomics to your spatial transcriptomic workflow by staining the same slide with a ready-to-go 40+-marker IMC protein panel after RNA detection. Learn how in 20 minutes.
Experiment considerations
For more detailed information on this workflow and how to plan your experiment, read the application note Combining Spatial Transcriptomics and Hyperion XTi Workflows for More Comprehensive Spatial Biology, which provides an overview of the application to detect protein targets using IMC analysis after transcript detection from the same slide.
With the versatility of IMC technology, the workflow allows any antibodies of interest to be added, or pre-configured and validated IMC panel sets to be used.
Visualize key RNA and protein markers in the same samples with spatial multi-omics
Spatial transcriptomics can be used in conjunction with other omics, such as spatial proteomics, to create a more complete picture of cell behavior and response. By combining spatial transcriptomics with spatial proteomics, knowledge of the cell’s transcriptome can improve understanding of cellular function and activation state, and cellular phenotypes can be detected using protein targets in the same tissue.
The complementary capabilities of spatial transcriptomics for transcript detection followed by use of the Hyperion XTi Imaging System for protein detection on the same slide enables high-quality data to be more directly assessed, providing a comprehensive understanding of tissue biology.
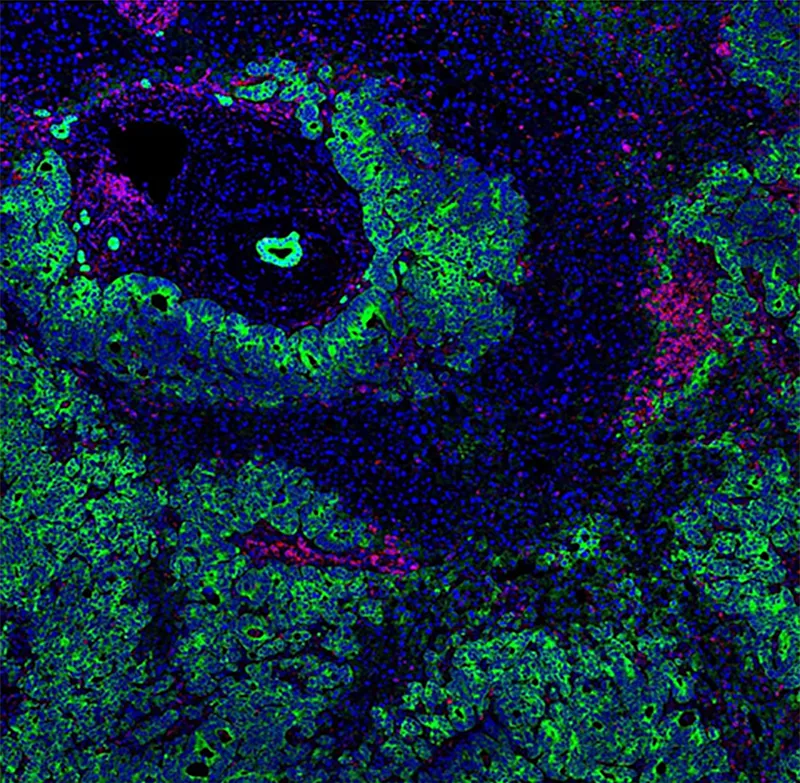
Hyperion systems: Ideal for combined spatial transcriptomic and spatial proteomic imaging
Assay development time: Because IMC technology uses an antibody reagent cocktail, it is simple to mix and match different antibodies without extensive assay validation.
Linear dynamic range and quantitation: Brightfield imaging (DAB) gives at best one order of magnitude (OoM) of dynamic range, while fluorescence gives 2–3 OoM and IMC technology gives 5 OoM. This enables the quantitation of weak signals in the presence of bright signals in the same sample.
Flexibility of tissue acquisition parameters (Hyperion imaging modes) and a 40-slide autoloader: Capture data from 40 slides in 24 hours, and run your whole study in an automated, walk-away run.
Resources – how scientists use spatial transcriptomics
Additional support: For support information and help with adapting the IMC protocol to your application, talk with one of our Field Applications Scientists here.
For Research Use Only. Not for use in diagnostic procedures. Patent and License Information: www.standardbio.com/legal/notices. Trademarks: www.standardbio.com/legal/trademarks. Any other trademarks are the sole property of their respective owners. ©2025 Standard BioTools Inc. All rights reserved.
