Targeted DNA-seq
Achieve economy of scale with ease
Reduce hands-on time and drive down reagent consumption with automated microfluidics, resulting in significant savings. And to ensure that your NGS panel always remains up-to-date, maintain flexible customization capabilities unique to Standard BioTools.

Juno targeted DNA sequencing library preparation system
Customized assay panels in convenient kits provide investigators and genomic service providers with an easily scalable and automated solution. Targeted DNA sequencing LP is compatible with Illumina MiniSeq, MiSeq, NextSeq and HiSeq NGS systems.
FEATURE | Advanta CFTR NGS Library Prep Assay saves time and money through an efficient and scalable targeted NGS workflow
The Advanta™ CFTR NGS Library Prep Assay automates targeted enrichment of cystic fibrosis transmembrane conductance regulator (CFTR) variants from each of the gene’s 27 exons and select intronic regions using proven microfluidics technology. The workflow significantly lowers hands-on time while conserving precious reagents by reducing reactions to the nanoliter scale. With the capacity to produce from 48 to over 1,500 NGS libraries per week, scalability is simplified to meet laboratory needs as they change over time. Protocols are available that support Juno™ and Access Array™ systems.
Methods
Juno targeted DNA sequencing LP
Scale with the industry’s most flexible per-sample throughput and enrichment capabilities using the Juno system. Two sample throughput options are available, the LP 48.48 IFC and the Juno LP 192.24 IFC (integrated fluidic circuit).
The LP 192.24 IFC automates amplicon-based libraries for 192 unique DNA samples per run and up to 2,400 amplicon targets per sample. The LP 48.48 IFC accommodates up to 48 DNA samples per run and up to 4,800 amplicon targets per sample. Targeted DNA sequencing LP is fully compatible with all Illumina sequencing systems. Up to 1,536 sample libraries may be multiplexed per sequencing run.
Access Array LP
Access Array delivers specimen flexibility supporting sample types of DNA extracted from germline and FFPE somatic samples. Access Array automates targeted DNA library preparation for 48 unique DNA samples per run and up to 4,800 amplicon targets per sample using Targeted DNA Seq Library reagents, or up to 480 amplicon targets per sample using Access Array chemistry.
The system is compatible with multiple sequencing platforms including Illumina sequencing systems and, for use with Access Array chemistry, Ion Torrent.
Advanta RNA-Seq XT NGS Library Prep Kits
Transform your RNA sequencing productivity with nanoscale automation
RNA sequencing (RNA-seq) is the standard for hypothesis-free profiling of the transcriptome and an essential tool for molecular biology laboratories. Designed to drive significant improvement in the RNA-seq workflow, the Advanta RNA-Seq NGS Library Prep Kit together with the Juno system provides an integrated solution for automated, cost-efficient NGS library prep.

We have developed an elegant microfluidics-based platform for automated RNA-seq library preparation. Leveraging the Juno system and the 48.Atlas™ IFC, the workflow supports the simultaneous processing of up to 48 total RNA samples with a one-click script on the instrument. With the launch of the Advanta RNA-Seq XT Library Prep Kits we have updated the products to include unique dual index (UDI) adapters, ideal for use on NovaSeq platforms, as well as simplified the reagent usage for additional savings on hands-on time and labor costs.
The Advanta RNA-Seq XT Kits contain sufficient 48.Atlas IFCs (four per kit), reagents, control line fluid and UDI adapters to generate libraries from 192 total RNA samples, and include either four sets of 1–48 UDI barcodes (PN 101-9950) or two sets of 1–48 UDI barcodes and two sets of 49–96 UDI barcodes (PN 101-9959).
The Advanta RNA-Seq XT NGS Library Prep Kits and Juno system advantage
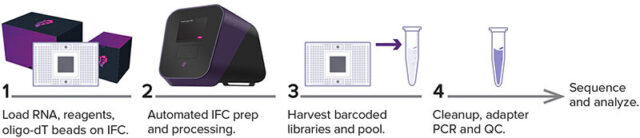
Figure 1. Advanta RNA-Seq NGS XT Library Prep Kit workflow on the Juno system. Samples and Advanta RNA-Seq XT reagents are added to the 48.Atlas IFC, which is subsequently processed on the Juno instrument.
The system solution automates many tedious, hands-on steps to generate full-length stranded libraries from random priming of polyadenylated [poly(A)] RNA present in total RNA samples. The Advanta RNA-Seq XT Kit and Juno workflow improve upon alternative RNA-seq library prep workflows by providing true walkaway automation while reducing reagent, consumables, labor and instrument costs.
WORKFLOW
Advanta RNA-Seq XT workflow, 48.Atlas IFC and the Juno platform enable significant efficiencies
The Advanta RNA-Seq XT Kits include library preparation reagents, UDI adapters and the 48.Atlas IFC, a microfluidic device approximately the same size and shape as a standard 96-well plate. Sample and reagent mixes are dispensed into the IFC, which is then placed in the Juno system for automated processing of most of the workflow steps. Because processing on the Juno platform requires no manual intervention and no pipetting, the solution greatly reduces hands-on time as well as consumable cost and plastic waste.
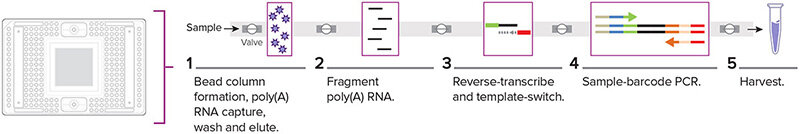
Figure 2. The 48.Atlas IFC architecture automates multiple workflow steps otherwise performed manually, including poly(A) RNA capture, RNA fragmentation, reverse-transcription, sample-barcode PCR and multiple wash steps.
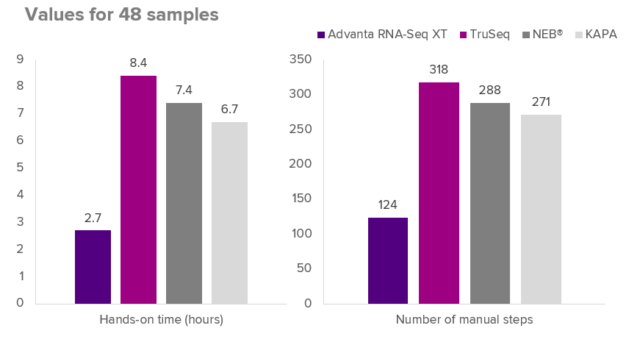
WORKFLOW COMPARISON
Figure 3. Example comparing hands-on time and number of manual processing steps for several RNA-seq workflows. Comparisons were made between the Advanta RNA-Seq XT NGS Library Prep Kit, TruSeq Stranded mRNA LP, NEBNext Ultra II Directional RNA LP and KAPA mRNA HyperPrep LP. All workflows include quantification of 48 samples by qPCR and pooling to obtain the final library for sequencing. The Advanta RNA-Seq XT workflow using the 48.Atlas IFC with Juno system automation enables substantial hands-on time savings relative to the alternative library prep workflows.
Performance
Summary of performance results generated from analytical validation testing.
Metric MeanUHRR
10ngUHRR
100ngBrain
10ngBrain
100ng
| Percent reads mapped to genome (GRCh38) | 94.7% | 95.1% | 94.7% | 94.5% |
| Percent reads mapped to transcriptome (RefSeq) | 77.7% | 75.8% | 73.1% | 68.3% |
| Percent rRNA reads | 1.5% | 2.2% | 3.7% | 5.7% |
| Percent correct strandedness | 97.7% | 98.1% | 97.6% | 98.1% |
Table 1. Performance characteristics of the Advanta RNA-Seq XT Kit on the Juno system were assessed in an analytical validation study. Total RNA samples were Universal Human Reference RNA (UHRR, Agilent) and human adult brain RNA (Brain, BioChain). The results demonstrate high mapping statistics with a low-percentage reads mapping to rRNA sequences for both RNA types and input amounts of either 10 ng or 100 ng.
Learn more: Download the Advanta RNA-Seq XT NGS Library Prep Kit Application Note.
Customer stories
Learn more about how our customers are leveraging Standard BioTools™ technology.